Views
A view is a type of Synapse Table that queries across metadata (Annotations) for particular items (currently: projects or files) with a particular “scope”. A File View lists all Files or Tables within one or more Folders or Projects. A Project View lists all Projects you’ve added to the view. Views can:
- Allow
Projects,Files, andTablesto be easily searched and queried - Allow view/editing metadata attributes in bulk
- Provide a way of isolating or linking data based on similarities
- Provide the ability to link
Projects,Files, andTablestogether by their annotations
Create a File View
To create a File View, select the Project in which you would like to create the view. The Project you choose does not have to contain the files you are including in your view. You will select the files of interest by defining the scope, which is the Project(s) and Folders that contain your files. “File Views” can also contain Tables or Folders; you can choose which kinds of items you would like to include during this process.
Instructions for creating Views using the clients can be found in the Python docs and in the R docs.
Create a Project View
To create a Project View, select the Project in which you would like to create the view. You will select the projects of interest by defining the scope as above. The only notable difference between creating a Project View and a File View is that for project views, there is a 1:1 relationship between the projects you select in your scope and the projects that are shown in the view.
Updating the Scope or Content-Type of a View
Views can be edited to change the scope of the view (e.g. the Project or Folder the view is showing) or which types of content is shown. Both of these options are found by navigating to “Show Scope of View” in the Tools menu; from there you may View and Edit the scope and the content-type of the view.
Note that it may take a few moments for the updated View to rebuild as it queries across the system.
Query a View
A view can be queried exactly the same as any other Table in Synapse. Please see Tables for more examples. See the Using Simple Search and Using Advanced Search sections below.
For example, to query for everything in syn123:
Command Line
synapse query 'SELECT * FROM syn123'
Python
query = syn.tableQuery('SELECT * FROM syn123')
R
query <- synTableQuery('SELECT * FROM syn123')
Update Annotations in Bulk
Views can be used to update annotations in bulk. To add new annotations, see the Annotations article. To update other metadata in bulk, such as provenance, see the Bulk Processing article.
For example, if you would like to use the Python client to update the annotation dogSays:bark to dogSays:woof in every file in a File View with the synId syn456, you can do:
from synapseclient import Table
foo = syn.tableQuery('select * from syn456')
bar = foo.asDataFrame()
# add in annotation as a column
bar['dogSays'] = 'woof'
# store the fileview with the new annotation in Synapse
fv = syn.store(synapseclient.Table(foo.tableId, bar))
Using Simple Search
Views are in Simple Search mode by default. You can filter out Projects or Files of interest by selecting what characteristics you like using the facet menu on the left. You can toggle between simple and advanced search using the Show advanced search/Show simple search link.
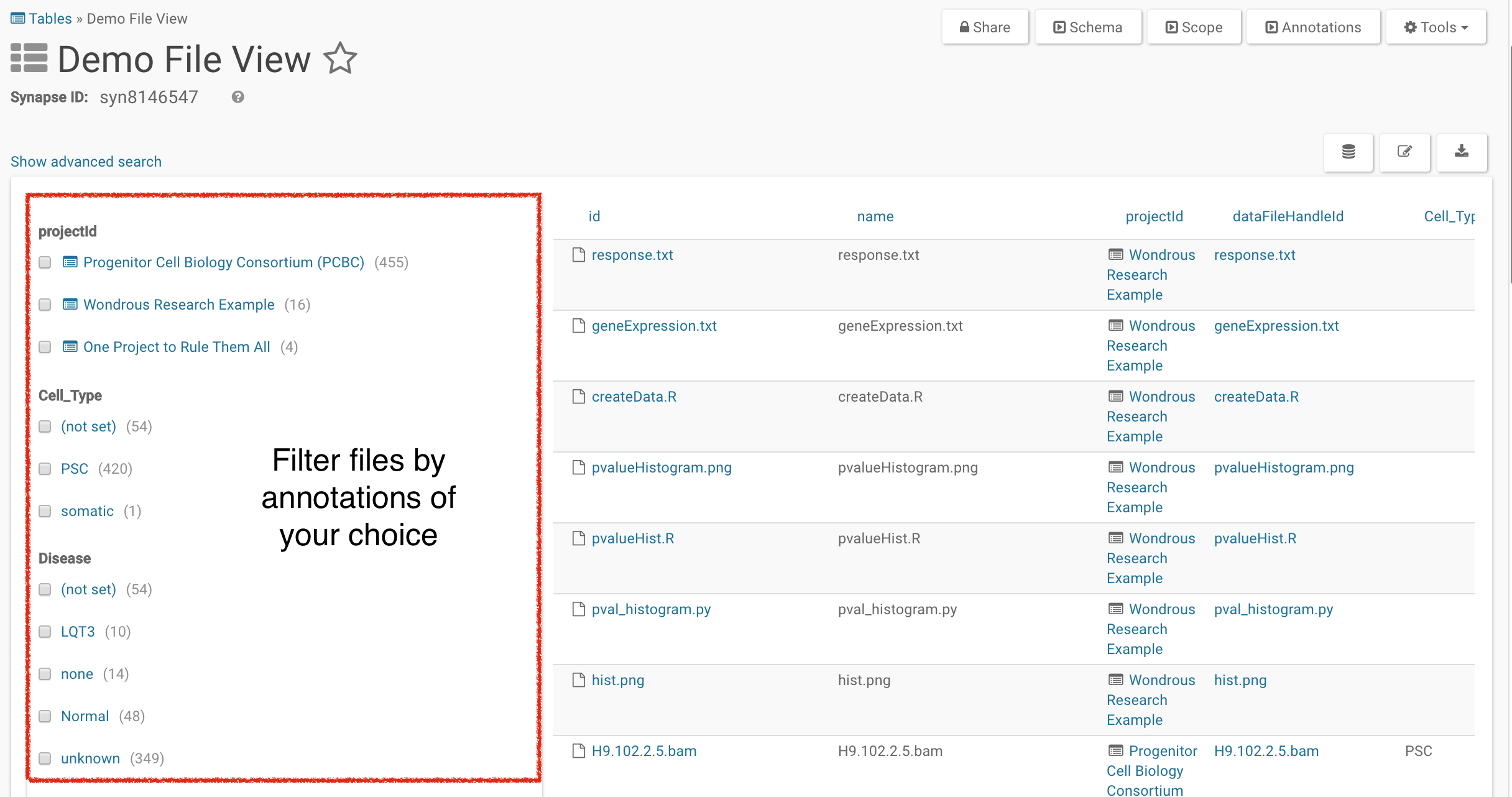
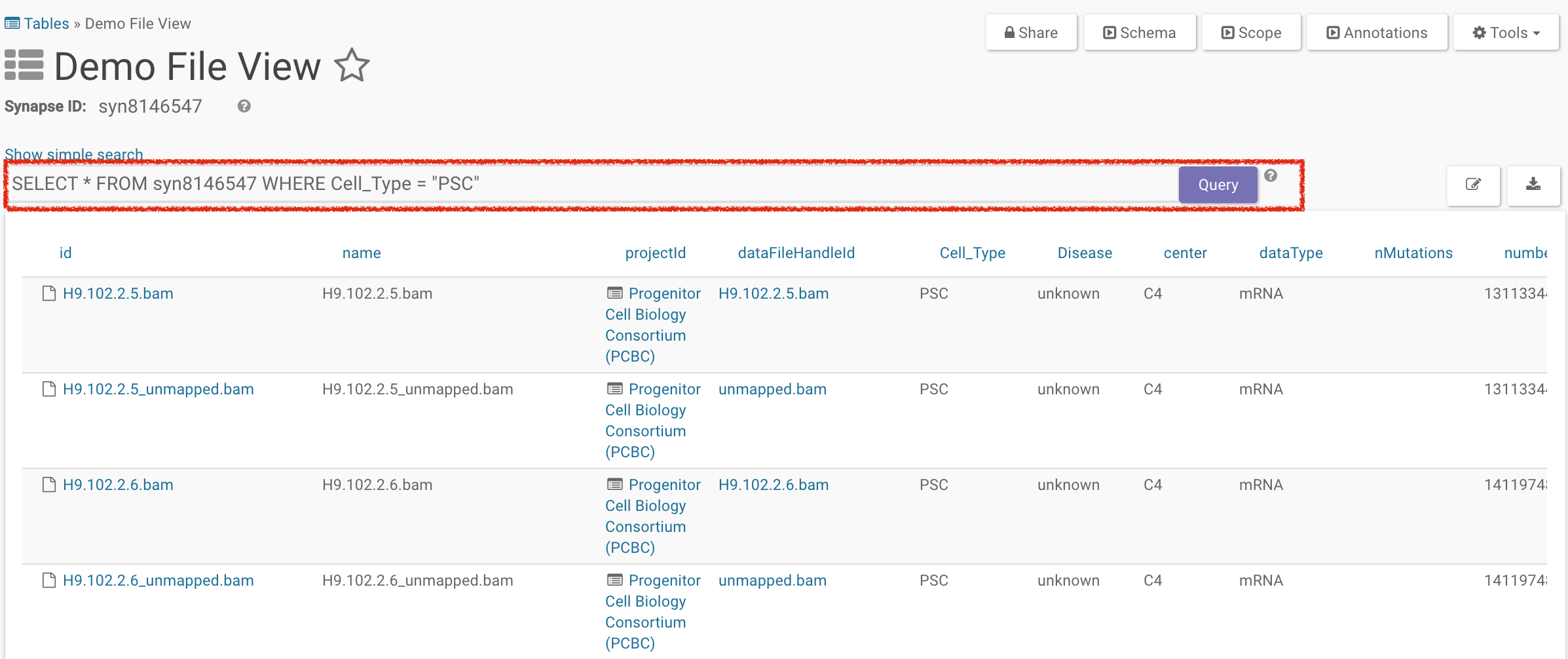
Using Advanced Search
In advanced search, you can use a SQL-like query to search for items in that view. In the example below, we’re selecting for all files that have a Cell Type of PSC.
Insert a View into a Wiki
Views can also be placed inside a Wiki once they have been created. You can embed the entire view or a subset of a query on it.
To insert a file view with a synId of syn8146547:
In the Edit Project Wiki window, select Table: Query on a Synapse Table/View under the Insert dropdown. To embed the entire file view into the wiki enter SELECT * FROM syn8146547 in the resulting pop-up.
To embed a subset of the file view, like the advanced search query in the previous example, enter SELECT * FROM syn8146547 WHERE Cell_Type = 'PSC'.
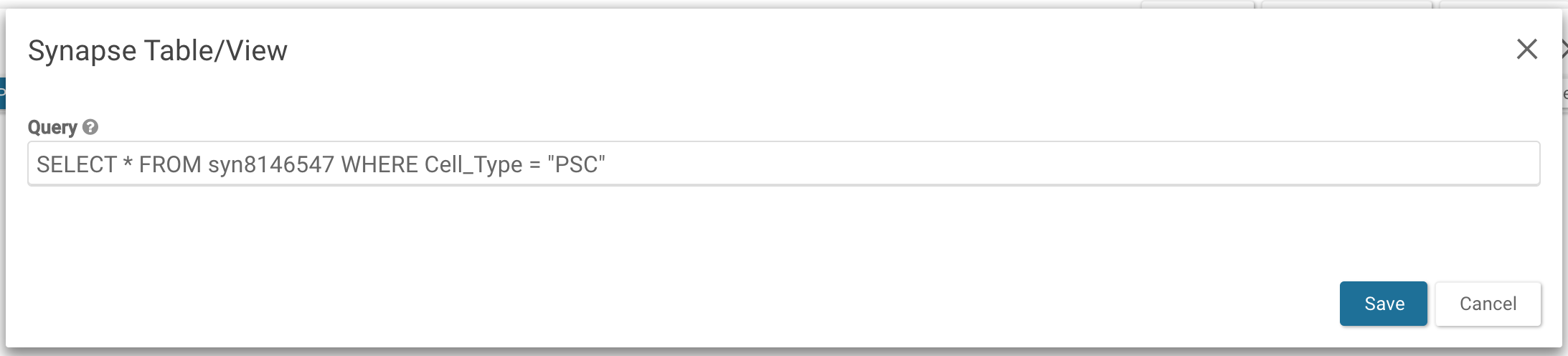
Save the query and the edits to the Wiki to embed the view.
See Also
Need More Help?
Try posting a question to our Forum.
Let us know what was unclear or what has not been covered. Reader feedback is key to making the documentation better, so please let us know or open an issue in our Github repository (Sage-Bionetworks/synapseDocs).