Using Tabular Data in Wikis
Use a Wiki Table Widget to Feature a Data Subset
Wiki widgets are tools to support narrative content. Embedded Wiki Tables can enable quick access to Files of highest importance and orient users to the resources contained in a collaborative project. Wiki Tables accept queries of existing Tables or File Views, providing a strategy to restrict scope and focus content.
More information on related concepts is available on the Tables, Making a Project, Views and Wikis pages.
Start with a File View or Table
The Views page provides steps to list Files and Folders within Views and Tables. This will be the content embedded in the Wiki.
Focus Scope
A Project can contain multiple studies, various assays and extensive Files, obscuring relevant data under a large file hierarchy. For this use case, an individual assay type is meant to be accessible from the Wiki, as the assay-relevant data is a subset of a broader study. The objective of highlighting .fastq and .bam rnaSeq files in an embedded Table is to provide an analyst a clear path to access files for reproducible studies.
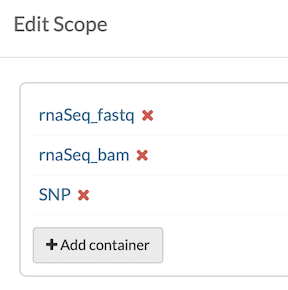
This study contains rnaSeq and SNP genomic data, as depicted in the fileview scope below. The scope can be further restricted to rnaSeq-specific data with additional queries on the embedded Table.
Restrict Query
A Table query is required to embed a Wiki Table. As the fileview is a combination of multiple assay types from a single study, the query must be restricted to isolate relevant rnaSeq entries. Additionally, columns visible can be limited to ensure only imperitive information is visible in the Wiki.
SELECT id,currentVersion,fileFormat,assay,genomeBuild,modifiedOn FROM syn17097374 WHERE "assay" = 'rnaSeq' AND "fileFormat" = 'bam' OR "fileFormat" = 'fastq'
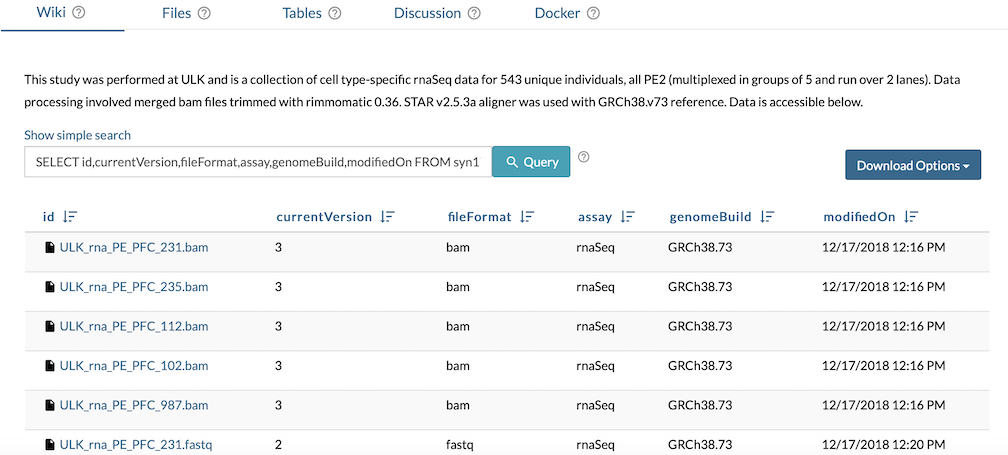
Embed Table
Navigate to Wiki Tools to Edit Project Wiki or to Folder Tools to Edit Folder Wiki. From the edit Wiki view, select Insert and Table:Query on a Synapse Table/View to insert the intended File View query. The result is a direct link to relevant files from a Wiki page with narrative content to provide additional context.
Need More Help?
Try posting a question to our Forum.
Let us know what was unclear or what has not been covered. Reader feedback is key to making the documentation better, so please let us know or open an issue in our Github repository (Sage-Bionetworks/synapseDocs).