Provenance
Reproducible research is a fundamental responsibility of scientists, but the best practices for achieving it are not established in computational biology. The Synapse “Provenance” system is one of many solutions you can use to make your work reproducible by you and others.
Provenance is a concept describing the origin of something; in Synapse it is used to describe the connections between workflow steps that derive a particular file of results. Data analysis often involves multiple steps to go from a raw data file to a finished analysis. Synapse’s Provenance Tools allow users to keep track of each step involved in an analysis, and share those steps with other users.
The basic elements of Synapse provenance
The model Synapse uses for provenance is based on the W3C provenance spec where items are derived from an activity which has components that were used and components that were executed. Think of the used items as input files and executed items as software or code. Both used and executed can either be items in Synapse or URLs such as a link to a github commit or a link to specific version of a software tool.
The Synapse clients for command line, Python, and R support creating and editing of provenance relationships. The Web client allows editing of provenance once the file has been uploaded.
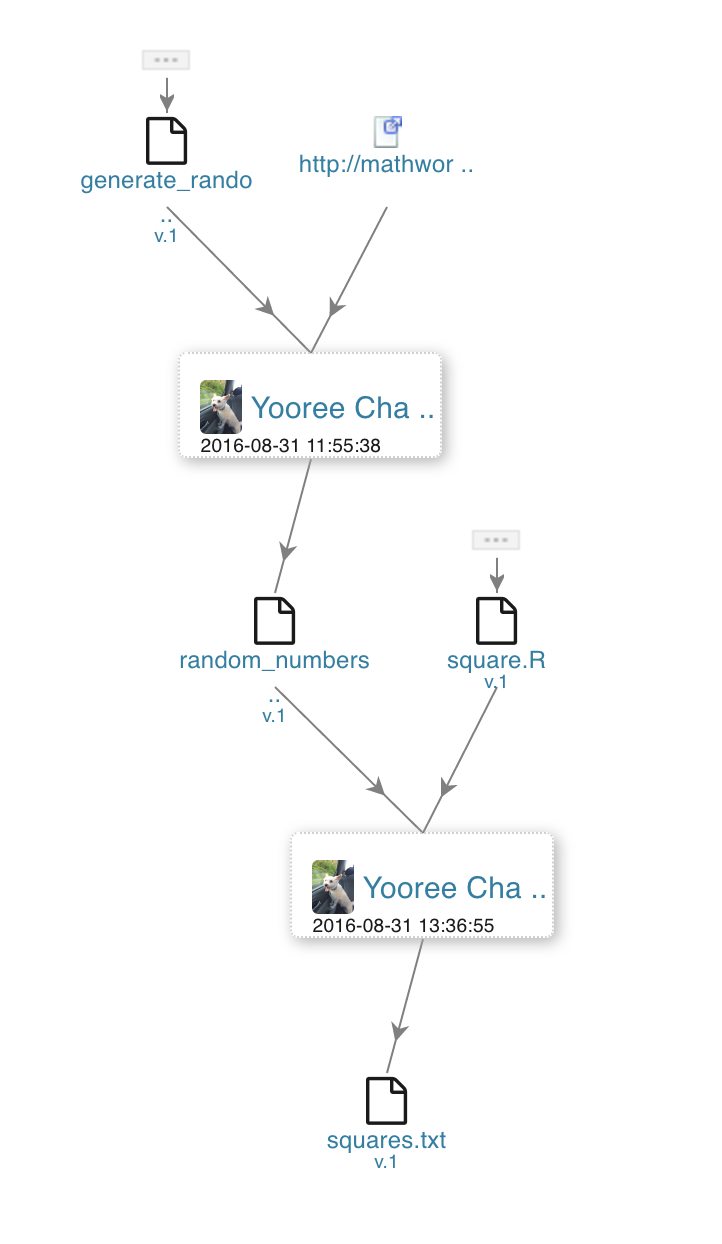
On the right is a Synapse visualization of provenance relationships that is demonstrated in the following section using our programmatic and web clients. In this example, we have two scripts, one that generates random numbers and another that takes a list of numbers and computes their squares. The project’s workflow looks like the image to the right.
Setting Provenance When Uploading a File
Let’s begin with a script that generates a list of normally distributed random numbers and saves the output to a file. For example, you have an R script file called generate_random_data.R and you’ve saved the output to a data file called random_numbers.txt. We’ll begin by uploading the files to Synapse and then set their provenance.
Upload a file and add provenance
For this example, we’ll use a Project that already exists (Wondrous Research Example : syn1901847). The code file is already saved in Synapse with synId syn7205215 so we’ll upload the data file to this Project, or in Synapse terminology, the project will be the parent of the new entities.
As the random_numbers.txt file was generated from the above script, we are going to specify this using provenance.
There are a couple ways to set provenance information for a Synapse entity. The used and executed arguments specify resources used and code executed in the process of creating the entity. Code can be stored in Synapse(as we did in the previous step) or, better yet, linked by URL to a source code versioning system like GitHub or SVN. As an example, we’ll specify 2 somewhat contrived sources of provenance:
- Synapse entity by synId: syn7205215 (the code file)
- URL to a page describing normal distributions
Web
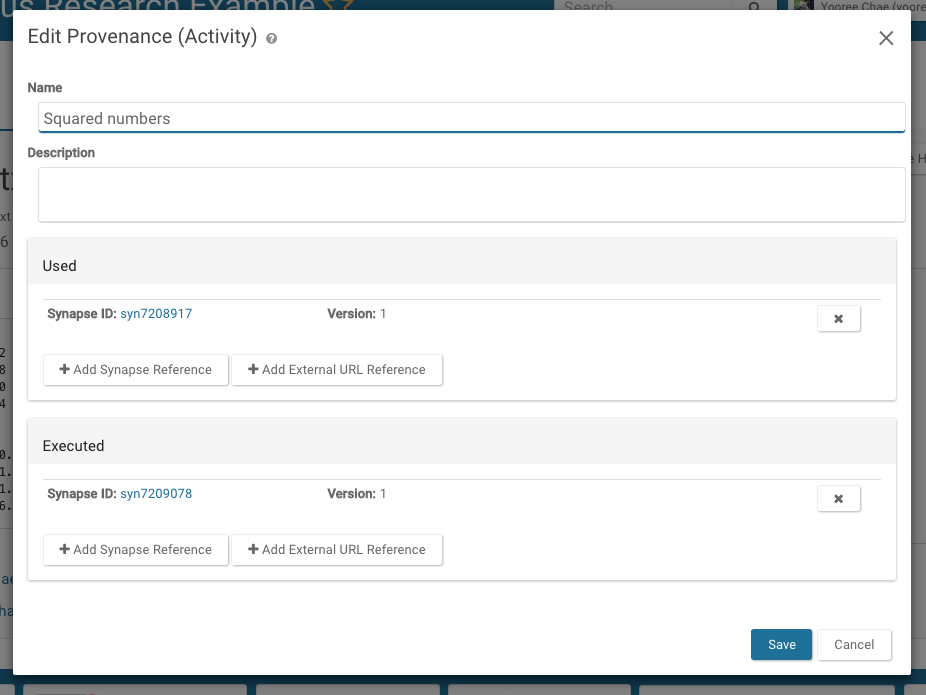
Navigate to the File's tab and click on the File that you would like to update. Click on the Tools dropdown in the upper right hand corner and select Edit Provenance. In the resulting pop-up, enter the relevant information.
Command Line
synapse add random_numbers.txt --parentId syn1901847 --executed syn7205215 --used http://mathworld.wolfram.com/NormalDistribution.html
Alternatively in the command line client, if you have downloaded the file, you can specify a local path as such:
synapse add random_numbers.txt --parentId syn1901847 --executed ./generate_random_data.R --used http://mathworld.wolfram.com/NormalDistribution.html
Python
# Set provenance for data file generated by the script file
data_file = File(path="random_numbers.txt", parent="syn1901847")
data_file = syn.store(data_file, executed="syn7205215", used="http://mathworld.wolfram.com/NormalDistribution.html")
R
# Set provenance for data file generated by the script file
data_file <- File(path="random_numbers.txt", parent="syn1901847")
data_file <- synStore(data_file, executed="syn7205215", used="http://mathworld.wolfram.com/NormalDistribution.html")
Once the data file is uploaded, it will provide the synId assigned to it. In this case, the data file’s synId is syn7208917.
Editing Provenance
To continue our example above, we’ll now add some new results from our initial data file. We’re going to take the results in random_numbers.txt and square them. The script to square the numbers will be square.R and we’ll save the output to a data file, squares.txt. As with the previous example, the code file is already saved in Synapse, so we’ll upload the data file and set its provenance.
Web
To update the provenance on a file, navigate to the File's tab and click on the File that you would like to update. Click on the Tools dropdown in the upper right hand corner and select Edit Provenance. In the resulting pop-up, enter the relevant information.
Command Line
# Add the data file to Synapse
synapse add squares.txt -parentId syn1901847
# Set the provenance for newly created entity syn7209166 using synId
synapse set-provenance -id syn7209166 -executed syn7209078 -used syn7208917
# Set the provenance for newly created entity syn7209166 using local path
synapse set-provenance -id syn7209166 -executed ./square.R -used ./random_numbers.txt
Python
# Add the data file to Synapse
squared_file = File(path="squares.txt", parentId="syn1901847")
squared_file = syn.store(squared_file)
# Set provenance for newly created entity syn7209166
squared_file = syn.setProvenance(squared_file, activity = Activity(used = "syn7208917", executed = "syn7209078"))
# Provenance can also be set using local variables instead of looking up synIds
squared_file = syn.setProvenance(squared_file, activity = Activity(used = data_file, executed = "syn7209078"))
R
# Add the data file to Synapse
squared_file <- File(path="squares.txt", parentId="syn1901847")
squared_file <- synStore(squared_file)
# Set provenance for newly created entity syn7209166
act <- Activity(name = "Squared numbers", used = "syn7208917", executed = "syn7209078")
synStore(squared_file, activity=act)
# Provenance can also be set using local variables instead of looking up synIds
act <- Activity(name = "Squared numbers", used = data_file, executed = "syn7209078")
squared_file <- synStore(squared_file, activity=act)
Deleting Provenance
If at any point you need to delete provenance on an entity, you can do so. You must be the person who created the entity to delete provenance.
Web
Navigate to the entity you would like to delete provenance from (e.g. a File or Folder). In this example, we are deleting provenance from a file. Select Tools->Edit File Provenance. In the list of Used and Executed, click the X to delete each activity and Save your changes.
Command Line
Currently, deleting provenance is not supported in the command line client.
Python
# Delete provenance on entity syn123
delete_provenance = syn.deleteProvenance('syn123')
R
# Delete provenance on entity syn123
deleteProvenance = synDeleteProvenance('syn123')
Getting and Viewing Provenance
Web
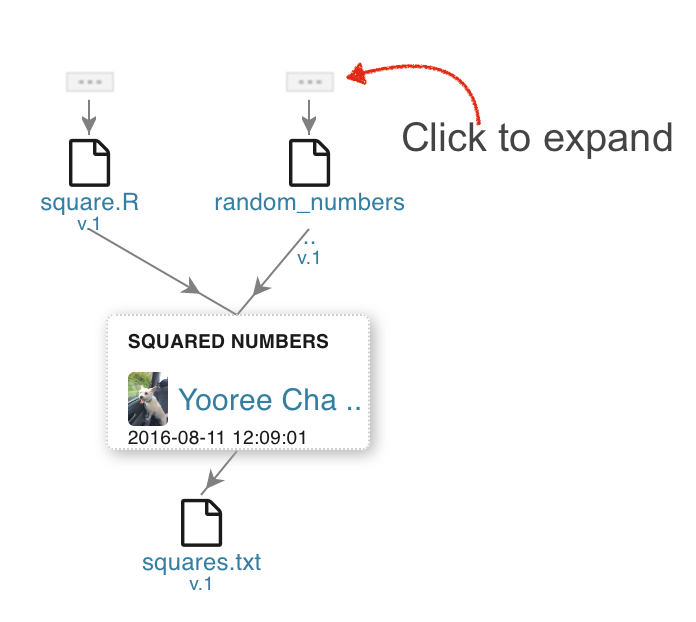
Navigate to the File's page to view its provenance. Clicking on the triple dots above entities will expand it to show the File's full provenance.
Command Line
synapse get-provenance -id syn7209166
Python
provenance = syn.getProvenance("syn7209166")
provenance
R
provenance <- synGetProvenance("syn7209166")
provenance
Reusing an Provenance for Multiple Files
An Activity is a Synapse object that helps keep track of what objects were ‘used’ in an analysis step … as well as what objects were generated. Thus, all relationships between Synapse objects and an Activity are governed by dependencies. That is, an Activity needs to know what it ‘used’ – and outputs need to know what Activity they were ‘generatedBy’. A couple of points for clarity:
- An
Activitycan ‘use’ many things (i.e. many inputs to an analysis) - Many outputs can be ‘generatedBy’ the same
Activity
If an activity isn’t assigned to an entity and then stored, a separate graph will be created for each file that the activity generated. The following example is used to assign the same activity to multiple files resulting in one provenance graph:
Web
Unfortunately, the web interface currently does not support assigning the same activity to multiple files.
Command Line
Unfortunately, command line currently does not support assigning the same activity to multiple files.
Python
# Code used to generate the file will be syn123456
# Files used to generate the information
expr_file = syn.get("syn246810", download=F)
filter_file = syn.get("syn135791", download=F)
# Activity to assign to multiple files
act = Activity(name="filtering",
used=[expr_file, filter_file],
executed="syn123456")
syn.store(final_file, activity=act)
# Get the activity now associated with an entity
act = syn.getProvenance(final_file)
# Now you can set this activity to as many files as you want (file1, file2, etc are Synapse Files)
file_list = [file_1, file_2, file_3]
file_list = map(lambda x: syn.store(x, activity=act), file_list)
R
# Code used to generate the file will be syn123456
# Files used to generate the information
expr_file <- synGet("syn246810", download=F)
filter_file <- synGet("syn135791", download=F)
# Activity to assign to multiple files
act <- Activity(name="filtering",
used=list(expr_file, filter_file),
executed="syn123456")
finalFile <- synStore(finalFile, activity=act)
# Get the activity now associated with an entity
act <- synGetProvenance(finalFile)
# Now you can set this activity to as many files as you want (file1, file2, etc are Synapse Files)
finalList <- c(file1, file2, file3)
finalList <- lapply(finalList, function(x) synStore(x, activity=act))
See Also
Need More Help?
Try posting a question to our Forum.
Let us know what was unclear or what has not been covered. Reader feedback is key to making the documentation better, so please let us know or open an issue in our Github repository (Sage-Bionetworks/synapseDocs).